In Helsinki Challenge, the Metabold-team focuses on the question what do the small molecules in our body, the metabolites, tell about our health and how do they affect it.
Small but so important metabolites
The human body contains several kinds of small molecules in different concentrations, even over one thousand different types of molecules can be found. Metabolites have many different roles, they carry energy, act as building blocks of cells, transmit messages within and between cells, and regulate how the cells operate.
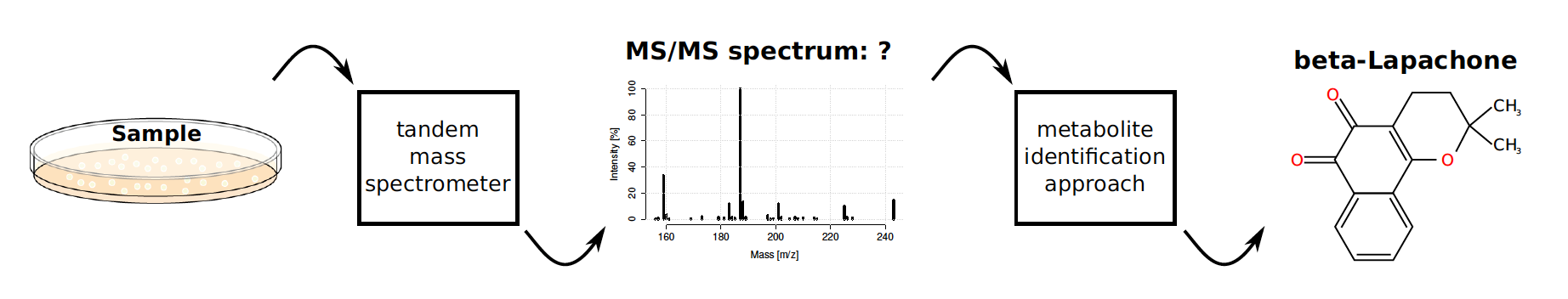
How small is small? For example glucose has a diameter of one nanometer (one billionth of a meter), 25000 times small and the width of a human hair and hundreds of times smaller than the wavelength of visible light. Small molecules can therefore not be seen or even measured in a simple way. Instead, one needs complex measurement devices and computational processing of the signal the devices output. The most sensitive method for observing small molecules is mass spectrometry (MS), which is based on separating ionized molecules in an electronic field based on their different behaviors.
It is not sufficient to measure the mass of the molecule to identify it, independently of how accurately one can measure. The reason is that there are a large number of alternative molecular structures. Even if one would know the molecular formula of the metabolite, in addition to the accurate mass, one may still have up to thousands of possible molecular structures. Metabolite identification is greatly helped by so called tandem mass spectrometry (MS/MS), that breaks the original molecule into fragments and then separates the fragments based on their masses. One molecule gives then a complete spectrum (MS/MS spectrum). Two molecules that have the same molecular formula but a different structure often break up into different groups of fragments, and their spectra then looks different.
Computers to the rescue in data crunching
A single sample of blood or saliva may result in a dataset of hundreds of tandem mass spectra, where one should identify the underlying metabolite. Sifting through this kind of data consumes both time and money. Luckily, computer- aided metabolite identification methods have developed rapidly in recent years. Predictive models can prune the set of candidate structures into a fraction of the original, which helps the work of the human expert and opens up new application possibilities.
The metabolite identification methods developed by the KEPACO research group of Aalto University (http://research.ics.aalto.fi/kepaco/) are based on machine learning. In machine learning, the computer is shown thousands of examples of MS/MS spectra and the corresponding molecular structures. Based on the examples the computer automatically builds a predictive model that associates the properties of the MS/MS spectrum to the properties of the molecular structure, for example the functional groups the molecule contains.
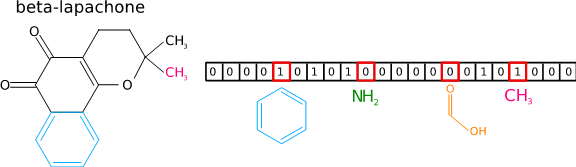
When the predictive model is shown a new spectrum, it will predict what kind of properties the corresponding molecule should have. Comparing the predicted properties to the alternative molecular structures one can finally construct a ranked list of candidate molecules, so that highly ranked molecules fit to the predicted molecular properties the best. The methods using this approach won the international CASMI-contest in 2016 (http://www.casmi-contest.org), where one searched for the most accurate metabolite identification methods.
Towards monitoring the metabolic health
Metabolite identification helps to understand how exercise, nutrition, alcohol use and diseases manifest in the metabolic balance. The goal of the Metabold team is indeed to develop metabolite monitoring as a service that may be used by doctors, athletes, self-monitoring enthusiasts as well as ‘regular’ consumers.
Juho Rousu
The author is a professor in Aalto University, whose reseach group develops (http://research.ics.aalto.fi/kepaco/) machine learning methods for small and slightly larger molecules. Pictures credit: Eric Bach
